
How have some species managed to colonize much of the planet? For more than a decade, we have been trying to answer this question for a globally-distributed bird, the house sparrow. Our current focus is to query whether some individuals can use their genomes adeptly to combat infections. When animals colonize new places, they must kill or control novel and familiar parasites, or the invasion fails. Our plan is to leverage the near-ubiquity of the house sparrow (Passer domesticus) to learn how the CpG (i.e., cytosines proximal to guanines in DNA sequences) content of gene regulatory regions influenced and is influencing the distribution of this avian species. CpG content, something we term epigenetic potential (EP), might allow genotypes to mask and manifest phenotypic plasticity reversibly within generations, as some CpG sites alter gene expression when methylated. We expect that EP represents a form of adaptive potential, providing organisms with a propensity to cope with the novel conditions. In particular, we expect EP to be important to the regulation of Toll-like receptors, major microbial surveillance mechanisms that reside on and in leukocytes. In a series of experiments and descriptive field and museum studies via an NSF grant, Aaron Schrey, Kevin Kohl, and the Martin lab are asking how are EP, DNA methylation, and TLR expression related within individual sparrows, whether EP for TLRs protective against gut microbial infections, if EP in TLRs influences gut microbial communities in ongoing range expansions, and whether EP for TLRs important to range expansions historically?
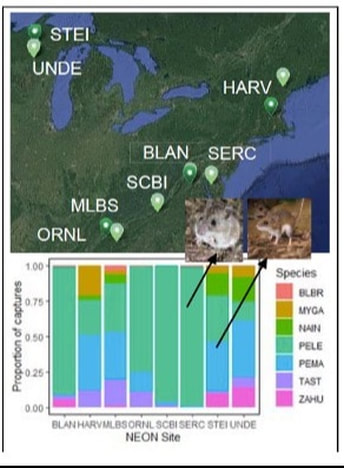
Many behavioral and immune system traits are sensitive to the environment, but the extent to which such variation drives infectious disease risk in nature remains obscure. For example, although it is known that particular rodent species are critical hosts of B. burgdorferi, the causative agent of Lyme disease, it is unknown how the environment affects traits that influence disease risk. This research project will leverage the continental scale of the National Ecological Observatory Network (NEON) to quantify behavioral and immunological traits of two keystone rodent hosts for B. burgdorferi, a pathogen that affects over 300,000 people in the U.S. each year. With NSF support, John Orrock and members of the Martin lab are testing the hypothesis that variation in the environment couples and decouples individual host traits, altering the abundance of competent hosts and driving the emergence of disease hotspots. This project will quantify variation B. burgdorferi competence in two rodent species across 8 NEON sites over 3 years and use statistical modeling to link individual variation in competence to variation in the abiotic and biotic environment and hence variation in Lyme disease risk. The utility of this approach will be further evaluated by using these models to forecast disease prevalence as a function of current climatic and habitat conditions at other NEON sites and in future years. Ultimately, the project will help us understand how organism-environment interactions drive the competence of individual hosts and will also help identify the level(s) of biological organization (e.g., individuals, populations, species) at which small changes have large consequences for disease risk.
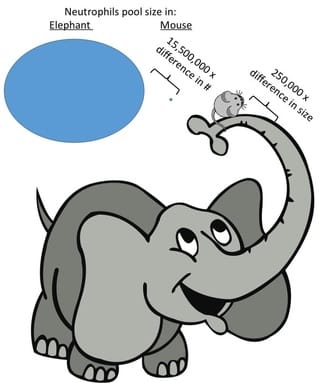
Body size affects almost all biological traits, but its influences on defenses, and especially immune systems, are largely unknown. This absence of information is surprising becuase large and small hosts likely experience quite different risks of parasite exposure and quite different consequences of infection. With funding from NSF and led by Cynthia Downs, we are asking how body size impacts the organization of innate immune defenses among terrestrial mammals spanning 7 orders of magnitude in size. We have already found that the circulating neutrophil pool is much larger than would be expected from isometry. Now, we're probing whether size affects antimicrobial activity in blood and the architecture of inflammation regulatory networks.
Other ongoing projects include:
1. Studying the role of FKBP5 and information in the regulation of glucocorticoids, steroid hormones animals use to cope with stressors, led by Cedric Zimmer;
2. Fostering interactions between organismal biologists and ecophysiologists with the National Ecological Observatory Network as part of an NSF RCN grant led by Ben Dantzer and with Karen Mabry;
1. Studying the role of FKBP5 and information in the regulation of glucocorticoids, steroid hormones animals use to cope with stressors, led by Cedric Zimmer;
2. Fostering interactions between organismal biologists and ecophysiologists with the National Ecological Observatory Network as part of an NSF RCN grant led by Ben Dantzer and with Karen Mabry;